Fundus Image Analysis
Jose Ignacio Orlando, Elena Prokofyeva and Matthew B. Blaschko
Overview:
In this work, we present an extensive description and evaluation of our method for blood vessel segmentation in fundus images based on a discriminatively trained, fully connected conditional random field model. Standard segmentation priors such as a Potts model or total variation usually fail when dealing with thin and elongated structures. We overcome this difficulty by using a conditional random field model with more expressive potentials, taking advantage of recent results enabling inference of fully connected models almost in real-time. Parameters of the method are learned automatically using a structured output support vector machine, a supervised technique widely used for structured prediction in a number of machine learning applications. Our method, trained with state of the art features, is evaluated both quantitatively and qualitatively on four publicly available data sets: DRIVE, STARE, CHASEDB1 and HRF. Additionally, a quantitative comparison with respect to other strategies is included. The experimental results show that this approach outperforms other techniques when evaluated in terms of sensitivity, F1-score, G-mean and Matthews correlation coefficient. Additionally, it was observed that the fully connected model is able to better distinguish the desired structures than the local neighborhood based approach. Results suggest that this method is suitable for the task of segmenting elongated structures, a feature that can be exploited to contribute with other medical and biological applications.
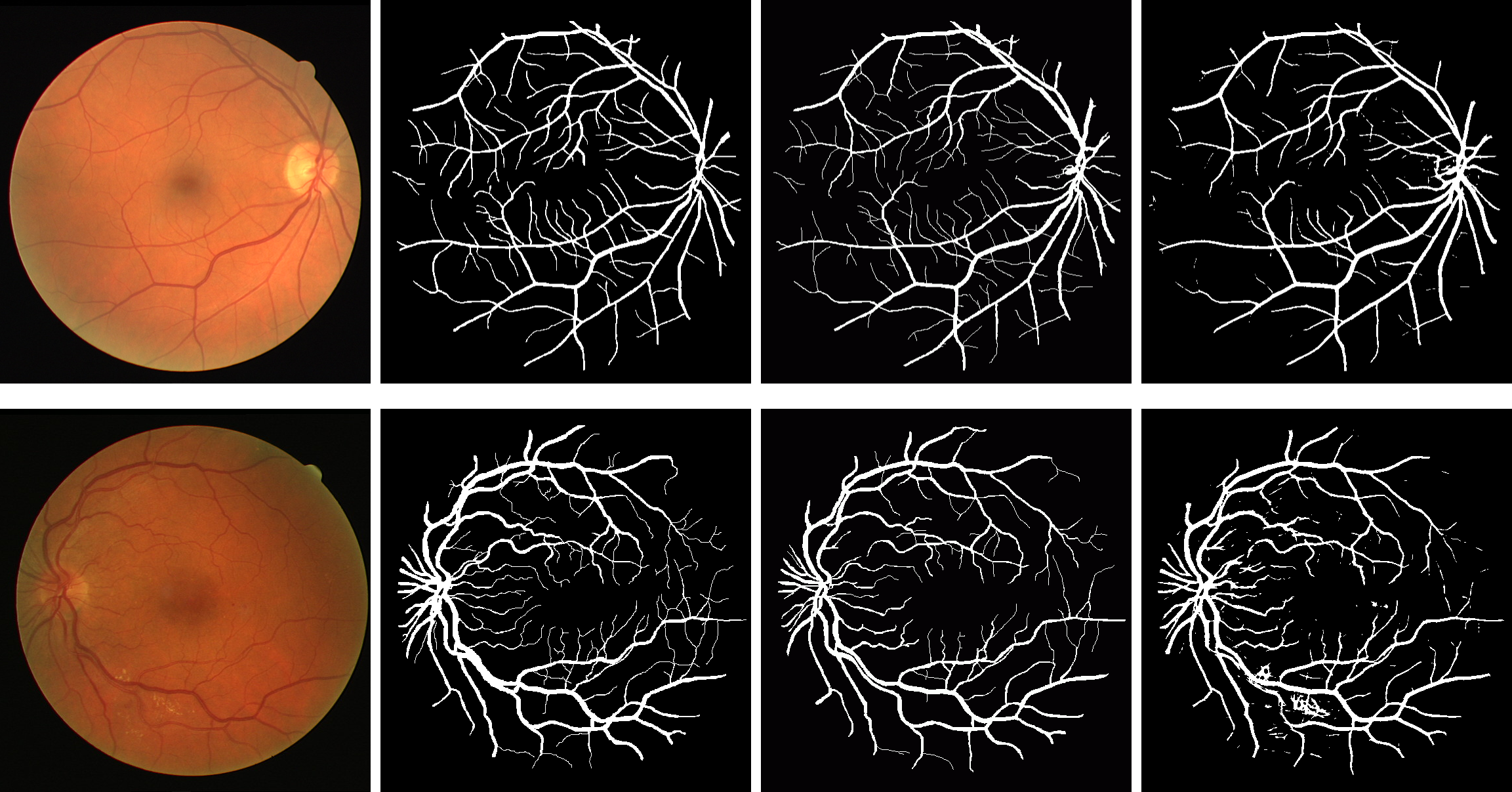
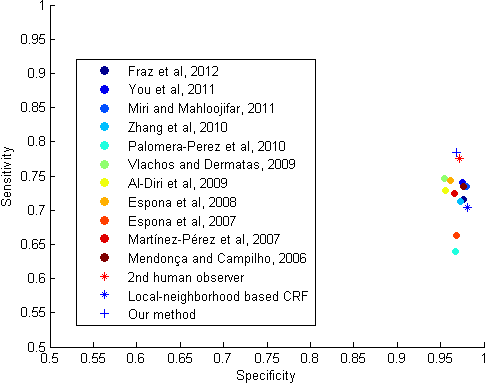
| Results obtained on healthy (top) and pathological (bottom) images from DRIVE (MICCAI 2014). | Scatter-plot of Se vs. Sp, comparing existing methods, local-neighborhood based CRF, the proposed method, and the human annotator, based on DRIVE data set (MICCAI 2014). |
 |  |
Code:
Code for segmenting retinal vessels using our TBME 2016 method is available here. If you use this software in your research, please cite the following papers:
Results:
MICCAI 2014 results:
TBME 2016 results:
Our segmentation method was evaluated on several data sets. They are available for download from the following webpages:
References: